Pan genome alignment tutorial
To quickly run the entire code and have all the input files, clone this repository.
git clone https://github.com/StevenVB12/Tutorial_pan_genomics
To make sure you have everything installed and setup, check here.
1. Introduction
In this tutorial we will align a piece of chromosome of two Heliconius butterfly species that includes the optix gene into a pan genome alignment.
Dorsal (top) and ventral (bottom) sides of Heliconius melpomene rosina (left) and Heliconius erato demophoon (right). The optix gene codes for a transcription factor that plays a key role in the development of red color pattern elements.

A pan genome refers to the complete set of genomic sequences shared by all individuals of a species, as well as the variable genomic sequences that are unique to specific individuals or subpopulations. We will use seq-seq-pan to construct the pan genome alignment, use some custom Python scripts to parse the outputs, and use R to visualize the alignment. Additionally, we will transform the coordinates of Transposable Element (TE) annotations and chromatin accessibility profiles (ATAC-seq) of developing head and wing tissues to the pan genome coordinate space and add them to this plot.
This tutorial follows up on the figure we built in the Minimap2 genome alignment tutorial.
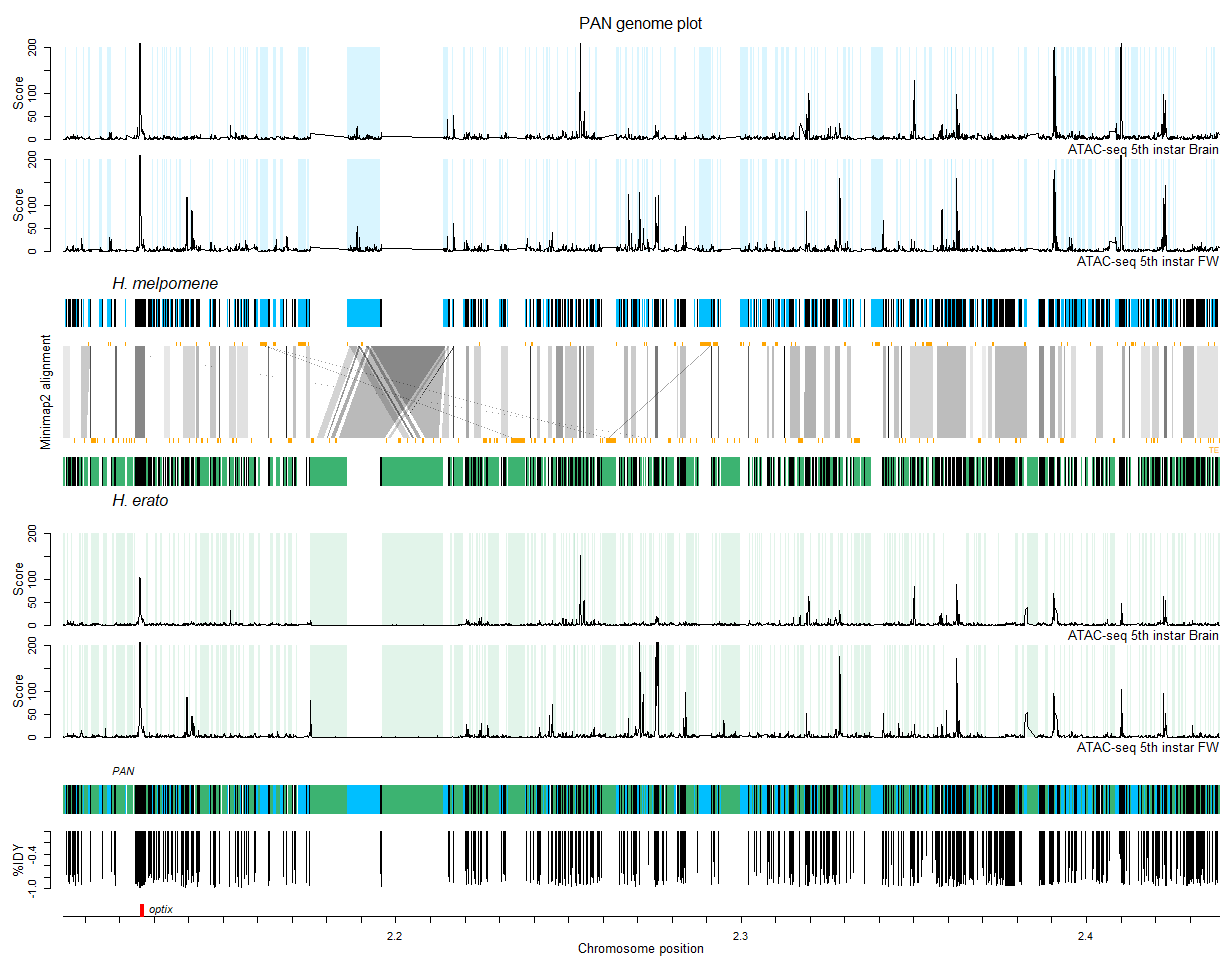
The final result should look like this:
Note: I will sometimes switch between code used in the Linux terminal or Rstudio. I've put these in black and blue boxes, respectively.
2. Input data
(This is the same as in the Minimap2 genome alignment tutorial)
For this exercise, you can navigate to http://ensembl.lepbase.org/ and click on the links to the Heliconius erato demophoon (v1) and Heliconius melpomene melpomene (Hmel2) genome assemblies. At the right top, you can search for "optix". The search result will return you the gene model name (e.g. evm.TU.Herato1801.64) and its location (e.g. scaffold 'Herato1801' at position '1239943' to '1251211').
# optix H. erato
Herato1801:1239943-1251211
# optix H. melpomene
Hmel218003:705604-706407
When you click on the gene model, you can see what genes surround the optix gene. You can also see a botton on the left "Export data". This allows you to export the sequence as a .fasta file. Using this, you can try exporting not just the optix gene but a 2,000,000 bp region surrounding it.
You can also find these .fasta files here.
# scaffold Herato1801 start 1 end 2000000
sequences/Herato1801_1_2000000.fasta
# scaffold Hmel213003 start 1 end 2000000
sequences/Hmel213003_1_2000000.fasta
3. seq-seq-pan aligner
We can use seq-seq-pan to assemble the sequences in the fasta files into a pan genome. Seq-seq-pan extends the functionality of the multiple genome aligner progressiveMauve by constructing a composite consensus or pan-genome that includes both homologous sequences or locally collinear blocks (LCBs) as well as lineage-specific (non-homologous) sequences in each of the genomes. This pan-genome is then used as the reference coordinates space for the multi genome alignment, which includes sequences specific to any of the genomes.
For our sequences, we will use seq-seq-pan as follows:
seq-seq-pan-wga --config genomefile=genome_list.txt outfilename=seq-seq-pan_out/SeqSeqPan_erato_melp_optix
The
genome_list.txtfile includes a list (one per line) of the fasta sequences to be included in the pan genome assembly. The file can be downloaded here.
Seq-seq-pan will output several files. Two will be relevenat for us here:
- The
_consensus.fastafile includes a complete fasta sequence of the consensus pan genome (stitching all non-homologous sequences into the assembly and taking the allele that is most frequent among the multiple aligned genomes). This consensus file demarks the coordinate space of the pan genome and will be used when we want to map any positions in the original genomes (e.g. TE positions) to the pan genome.- The
.xmfafile includes a list of the locally collinear blocks (LCBs). We will use this file to identify the sequences that are homologous or species-specific.> 1:sequence identifier of the first genome in thegenome_list.txtfile.> 2:sequence identifier of the second genome in thegenome_list.txtfile. (and so on)=demarks separate LCBs.-gaps in the aligned LCBs.
That's it, we have a pan genome!
4. Shared and unique sequences
We can now try to identify what parts of the sequences is identified as homologous or species-specific in the pan genome. We will use a custom Python script for this, available here.
# For the Python script to work, we first need to remove newlines (enters) in the file from lines that include the sequence. This can be done with this perl oneliner:
perl -pe 'chomp if /^[ATCGNSBDHVMRWYK-]/' seq-seq-pan_out/SeqSeqPan_erato_melp_optix.xmfa| sed 's/\=/\n\=/g' | sed 's/>/\n>/g' | sed '/^$/d' > seq-seq-pan_out/SeqSeqPan_erato_melp_optix.noNewline.xmfa
# Now we can run the python script ('-g 1,2' is a list of the genome identifiers in the order of the genomes_list.txt file):
python seq-seq-pan_blocks_intervals.py -I seq-seq-pan_out/SeqSeqPan_erato_melp_optix.noNewline.xmfa -g 1,2
# The script annoyingly produces a 'TAB' at the end of each line which would break bedtools in the next step. We can remove that as follows:
sed -i 's/[[:space:]]*$//' seq-seq-pan_out/1_blocks_intervals.bed
sed -i 's/[[:space:]]*$//' seq-seq-pan_out/2_blocks_intervals.bed
The output are files with
| chromosome | start | end |positions of sequences in each genome but in the coordinate space of the pan genome (hence, a new line is generate when teh sequence is interrupted by a species-specific sequence in another genome). Such a format with start and end positions is typically called a.bedfile format.
Next, we can use bedtools to subtract these files and get the unique sequences in each genome.
bedtools subtract -sorted -a 1_blocks_intervals.bed -b 2_blocks_intervals.bed > blocks_unique_1.bed
bedtools subtract -sorted -b 1_blocks_intervals.bed -a 2_blocks_intervals.bed > blocks_unique_2.bed
We also have a custom Python scripts to calculate sequence identity within the LCBs, available here.
# First, we need to transform the .xmfa file to a .fasta alignment
python seq-seq-pan_toFasta.py -I seq-seq-pan_out/SeqSeqPan_erato_melp_optix.noNewline.xmfa -g 1,2
cat genome* > seq-seq-pan_out/SeqSeqPan_erato_melp_optix.fasta
# Next, we need to find the sequences that are shared between genomes. We can do this with bedtools intersect.
bedtools intersect -sorted -a seq-seq-pan_out/1_blocks_intervals.bed -b seq-seq-pan_out/2_blocks_intervals.bed > seq-seq-pan_out/blocks_shared_1_2.bed
# Finally, we can calculate the conservation. This will output a .bed like file with identity scores for each shared interval.
python seq-seq-pan_bedfile_conservation.py -I seq-seq-pan_out/SeqSeqPan_erato_melp_optix.fasta -g 1,2 -b seq-seq-pan_out/blocks_shared_1_2.bed -o seq-seq-pan_out/conservation_shared_1_2.bed
5. Mapping annotations to the pan genome
The map function of seq-seq-pan allows to transform any original position of the included genomes to the pan genome (= the pan genome coordinates). The function takes as input a file with a single column of positions and a first row that specifies from where and to where to map (e.g. 2\tc, which means map from genome 2 (the Hmel218003 sequence which was the second genome in the genome_list.txt file) to genome c (the consesus pan genome sequence)).
Here I have the start and end position of the optix gene on the Hmel218003 scaffold (you could also get these or the positions of any gene from http://ensembl.lepbase.org/.
2 c
705604
706407
To run the mapping function:
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i gene_annotation/optix_Hmel2.toMap.txt -n gene_annotation/optix_Hmel2.toMap.pan
Let's plot the pan genome in R now. Later We'll use the map function to transform the positions of TEs and ATAC-seq bedgraph files.
6. Visualizations in R
Now we can switch again to Rstudio (but we will be generating some extra input files in Linux throughout the pipeline).
6.1. Set you working directory:
setwd("G:/My Drive/Workshop_pan_genomics_12042023")
(You can also use the '...' under the 'File' tab on the right of Rstudio to navigate to your folder then click 'More' > 'Set as working directory'.
6.2. Load data obtained from the seq-seq-pan genome
# Load the blocks that are unique to each genome
unique_erato <- read.table('seq-seq-pan_out/blocks_unique_1.bed')
unique_melp <- read.table('seq-seq-pan_out/blocks_unique_2.bed')
shared <- read.table('seq-seq-pan_out/blocks_shared_1_2.bed')
# set the column names
colnames(unique_erato) <- c( 'scaffold','startPos','endPos')
colnames(unique_melp) <- c( 'scaffold','startPos','endPos')
colnames(shared) <- c( 'scaffold','startPos','endPos')
# We ca filter out smaller unique blocks to not overload R (and visually even very small blacks will become plotted having a length of at least one pixel at zoomed out scales, which might not be correct).
unique_erato <- subset(unique_erato, abs(unique_erato$endPos - unique_erato$startPos) > 10)
unique_melp <- subset(unique_melp, abs(unique_melp$endPos - unique_melp$startPos) > 10)
# Load the sequence identity data of the blocks that are shared between species.
conservarion <- read.table('seq-seq-pan_out/conservation_shared_1_2.bed.txt', header = TRUE)
6.3. Build the layout of the plot.
Here, we'll build a layout with a total of 12 panels that we will fill with data (inlcuding one for the title, x-axis labels, ATAC-sea data, genomes, pan genomes, Minimap2 alignment, TEs, sequence identity, and the optix gene annotation). For more details on the layout() function please see the section '4.6. Zoom in' of the Minimap2 genome alignment tutorial.
layout(matrix(c(1,8,9,3,12,4,10,11,5,6,7,2), nrow=12, byrow=TRUE), height = c(0.1,0.3,0.3,0.2,0.3,0.2,0.3,0.3,0.2,0.2,0.07,0.1))
layout.show(n=12)
par(mar = c(0.5,5,0.5,1), xpd = FALSE)
# Set the zoom window for the plot
# This is for the entire pan genome
start = 0
end = 3124353 # This is the length of the pan genome. We got this value from running the script seq-seq-pan_blocks_intervals.py
# This is for a subwindow centered on optix
start = 2126065 -10000
end = 2126065 +300000
6.4. Plot the genomes in the pan genome space
# Plot the title of the plot
plot(NULL, xlim=c(start,end), ylim = c(0,1), axes=FALSE, ann=FALSE)
mtext('PAN genome plot', side = 1, cex=1, col = 'black', line =-1)
# Plot the x-axis as a separate plot
plot(NULL, xlim=c(start,end), ylim = c(0,1), axes=FALSE, ann=FALSE)
axis(1, at = seq(0,3124353, by=10000), labels = NA, line =-3)
axis(1, at = seq(0,3124353, by=100000), labels = round(seq(0/1000000,3124353/1000000, by=0.1),1), line =-3)
mtext('Chromosome position', side = 1, cex=0.8, col = 'black', line =-1)
## Plot H. melpomene genome plus the unique parts.
plot(NULL, xlim = c(start, end), ylim = c(0,10), axes=F, ylab = '', xlab = '')
text(start, 7.5, substitute(paste(italic('H. melpomene'))), pos = 4, cex = 1.5)
# Plot the shared blocks
for(e in 1: nrow(shared)){
rect(shared$startPos[e],0,shared$endPos[e],5, col = 'black', border = NA)
}
# Plot the unique blocks
for(e in 1: nrow(unique_melp)){
rect(unique_melp$startPos[e],0,unique_melp$endPos[e],5, col = 'deepskyblue', border = NA)
}
## Plot H. erato genome plus the unique parts.
plot(NULL, xlim = c(start, end), ylim = c(0,10), axes=F, ylab = '', xlab = '')
text(start, 2.5, substitute(paste(italic('H. erato'))), pos = 4, cex = 1.5)
# Plot the shared blocks
for(e in 1: nrow(shared)){
rect(shared$startPos[e],5,shared$endPos[e],10, col = 'black', border = NA)
}
# Plot the unique blocks
for(e in 1: nrow(unique_erato)){
rect(unique_erato$startPos[e],5,unique_erato$endPos[e],10, col = 'mediumseagreen', border = NA)
}
# Plot the whole pan genome.
plot(NULL, xlim = c(start, end), ylim = c(0,10), axes=F, ylab = '', xlab = '')
# rect(0,0,3124353,5, col = 'black', border = NA)
text(start, 7.5, substitute(paste(italic('PAN'))), pos = 4)
# Plot the shared blocks
for(e in 1: nrow(shared)){
rect(shared$startPos[e],0,shared$endPos[e],5, col = 'black', border = NA)
}
# Plot the unique blocks H. melpomene
for(e in 1: nrow(unique_melp)){
rect(unique_melp$startPos[e],0,unique_melp$endPos[e],5, col = 'deepskyblue', border = NA)
}
# Plot the unique blocks H. H. erato
for(e in 1: nrow(unique_erato)){
rect(unique_erato$startPos[e],0,unique_erato$endPos[e],5, col = 'mediumseagreen', border = NA)
}
# Plot conservation
plot(NULL, xlim = c(start, end), ylim = c(-1,0), axes=F, ylab = '', xlab = '')
for(e in 1: nrow(conservarion)){
rect(conservarion$start[e],0,conservarion$end[e],-conservarion$IDY[e], col = 'black', border = NA)
}
# Add a y-axis to the conservation plot
axis(2, at = seq(-1,0, by=0.2), line = 1)
mtext('%IDY', side = 2, cex=0.8, line = 3)
The genomes of both species as well as the composite pan genome should now be plotted with the unique blocks colored in blue and green and the shared blocks in black.
We should have the tranformed positions of the optix gene (see section '5. Mapping annotations to the pan genome') and we can add these to the plot:
# Plot optix gene annotation
plot(NULL, xlim = c(start, end), ylim = c(0,10), axes=F, ylab = '', xlab = '')
rect(2126065,0,2126871,10, col = 'red', border = 'red') # position transferred from optix melpomene (only has one exon)
text(2126871, 5, substitute(paste(italic('optix'))), pos = 4)
6.5. Map the ATAC-seq data to the pan genome and plot ATAC
6.5.1. Create ATAC-seq mapping files
Here we will use R to read in the ATAC-seq bedgraphs and create the input files for the seq-seq-pan map function. The transformed positions will in a later step be merged with the ATAC-seq read count data in the original files.
# Read in the ATAC-seq bedgraph files.
erato_5th_brain <- import.bedGraph("ATAC/brain_5th_H_erato_normalized_mean.w30s0bin.bg")
erato_5th_FW <- import.bedGraph("ATAC/FW_5th_H_erato_normalized_mean.w30s0bin.bg")
melp_5th_brain <- import.bedGraph("ATAC/brain_5th_H_melp_normalized_mean.w30s0bin.bg")
melp_5th_FW <- import.bedGraph("ATAC/FW_5th_H_melp_normalized_mean.w30s0bin.bg")
# Extract the start and end position of the bedgraph files.
erato_5th_brain_start <- as.data.frame(start(erato_5th_brain))
erato_5th_brain_end <- as.data.frame(end(erato_5th_brain))
erato_5th_FW_start <- as.data.frame(start(erato_5th_FW))
erato_5th_FW_end <- as.data.frame(end(erato_5th_FW))
melp_5th_brain_start <- as.data.frame(start(melp_5th_brain))
melp_5th_brain_end <- as.data.frame(end(melp_5th_brain))
melp_5th_FW_start <- as.data.frame(start(melp_5th_FW))
melp_5th_FW_end <- as.data.frame(end(melp_5th_FW))
# Add the first row which specifies the tranform direction.
colnames(erato_5th_brain_start) <- paste("1\tc")
colnames(erato_5th_brain_end) <- paste("1\tc")
colnames(erato_5th_FW_start) <- paste("1\tc")
colnames(erato_5th_FW_end) <- paste("1\tc")
colnames(melp_5th_brain_start) <- paste("2\tc")
colnames(melp_5th_brain_end) <- paste("2\tc")
colnames(melp_5th_FW_start) <- paste("2\tc")
colnames(melp_5th_FW_end) <- paste("2\tc")
# Write the objects to files.
write.table(erato_5th_brain_start, "ATAC/erato_5th_brain_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(erato_5th_brain_end, "ATAC/erato_5th_brain_end.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(erato_5th_FW_start, "ATAC/erato_5th_FW_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(erato_5th_FW_end, "ATAC/erato_5th_FW_end.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(melp_5th_brain_start, "ATAC/melp_5th_brain_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(melp_5th_brain_end, "ATAC/melp_5th_brain_end.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(melp_5th_FW_start, "ATAC/melp_5th_FW_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(melp_5th_FW_end, "ATAC/melp_5th_FW_end.toMap.txt", quote = FALSE, row.names = FALSE)
Note: For bigger files, you could also use the script seq-seq-pan_bedgraph_chrompos.py provided here which extracts the columns of the bedgrap file. The script also takes a chromosome position file which is able to generate cummulative chromosome positions. This is because seq-seq-pan generates the pan genome as a single string/chromosome when provided multiple chromosomes.
6.5.2. Transform the ATAC-seq positions
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i ATAC/erato_5th_brain_start.toMap.txt -n ATAC/erato_5th_brain_start.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i ATAC/erato_5th_brain_end.toMap.txt -n ATAC/erato_5th_brain_end.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i ATAC/erato_5th_FW_start.toMap.txt -n ATAC/erato_5th_FW_start.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i ATAC/erato_5th_FW_end.toMap.txt -n ATAC/erato_5th_FW_end.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i ATAC/melp_5th_brain_start.toMap.txt -n ATAC/melp_5th_brain_start.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i ATAC/melp_5th_brain_end.toMap.txt -n ATAC/melp_5th_brain_end.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i ATAC/melp_5th_FW_start.toMap.txt -n ATAC/melp_5th_FW_start.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i ATAC/melp_5th_FW_end.toMap.txt -n ATAC/melp_5th_FW_end.pan
6.5.3. Read and plot the transformed ATAC positions
erato_5th_brain_panPos_start <- read.table('ATAC/erato_5th_brain_start.pan.txt', header = TRUE, sep = '\t')
erato_5th_brain_panPos_end <- read.table('ATAC/erato_5th_brain_end.pan.txt', header = TRUE, sep = '\t')
erato_5th_FW_panPos_start <- read.table('ATAC/erato_5th_FW_start.pan.txt', header = TRUE, sep = '\t')
erato_5th_FW_panPos_end <- read.table('ATAC/erato_5th_FW_end.pan.txt', header = TRUE, sep = '\t')
melp_5th_brain_panPos_start <- read.table('ATAC/melp_5th_brain_start.pan.txt', header = TRUE, sep = '\t')
melp_5th_brain_panPos_end <- read.table('ATAC/melp_5th_brain_end.pan.txt', header = TRUE, sep = '\t')
melp_5th_FW_panPos_start <- read.table('ATAC/melp_5th_FW_start.pan.txt', header = TRUE, sep = '\t')
melp_5th_FW_panPos_end <- read.table('ATAC/melp_5th_FW_end.pan.txt', header = TRUE, sep = '\t')
# Merge the old with the newly transformed positions.
erato_5th_brain_PAN <- cbind(as.data.frame(erato_5th_brain), erato_5th_brain_panPos_start[,2], erato_5th_brain_panPos_end[,2])
erato_5th_FW_PAN <- cbind(as.data.frame(erato_5th_FW), erato_5th_FW_panPos_start[,2], erato_5th_FW_panPos_end[,2])
melp_5th_brain_PAN <- cbind(as.data.frame(melp_5th_brain), melp_5th_brain_panPos_start[,2], melp_5th_brain_panPos_end[,2])
melp_5th_FW_PAN <- cbind(as.data.frame(melp_5th_FW), melp_5th_FW_panPos_start[,2], melp_5th_FW_panPos_end[,2])
# Add readable column names.
colnames(erato_5th_brain_PAN) <- c('seqnames', 'start', 'end', 'width', 'strand', 'score' , 'panStart', 'panEnd')
colnames(erato_5th_FW_PAN) <- c('seqnames', 'start', 'end', 'width', 'strand', 'score' , 'panStart', 'panEnd')
colnames(melp_5th_brain_PAN) <- c('seqnames', 'start', 'end', 'width', 'strand', 'score' , 'panStart', 'panEnd')
colnames(melp_5th_FW_PAN) <- c('seqnames', 'start', 'end', 'width', 'strand', 'score' , 'panStart', 'panEnd')
# Remove potential erroneous mappings which can be identified as start/end positions too far apart.
erato_5th_brain_PAN <- subset(erato_5th_brain_PAN, abs(erato_5th_brain_PAN$panEnd - erato_5th_brain_PAN$panStart) < 10000)
erato_5th_FW_PAN <- subset(erato_5th_FW_PAN, abs(erato_5th_FW_PAN$panEnd - erato_5th_FW_PAN$panStart) < 10000)
melp_5th_brain_PAN <- subset(melp_5th_brain_PAN, abs(melp_5th_brain_PAN$panEnd - melp_5th_brain_PAN$panStart) < 1000)
melp_5th_FW_PAN <- subset(melp_5th_FW_PAN, abs(melp_5th_FW_PAN$panEnd - melp_5th_FW_PAN$panStart) < 1000)
# Sort the tables (this is needed because when plotting lines x coordinates have to be ordered)
erato_5th_brain_PAN <- erato_5th_brain_PAN[order(erato_5th_brain_PAN$panStart),]
erato_5th_FW_PAN <- erato_5th_FW_PAN[order(erato_5th_FW_PAN$panStart),]
melp_5th_brain_PAN <- melp_5th_brain_PAN[order(melp_5th_brain_PAN$panStart),]
melp_5th_FW_PAN <- melp_5th_FW_PAN[order(melp_5th_FW_PAN$panStart),]
## Finally, let's start plotting the ATAC-seq tracks
## Plot H. melpomene brain
plot(NULL, xlim=c(start,end), ylim = c(0,200), axes=FALSE, ann=FALSE)
# Add some shading for unique blocks under the plot
for(e in 1: nrow(unique_melp)){
rect(unique_melp$startPos[e],0,unique_melp$endPos[e],200, col = adjustcolor('deepskyblue',alpha.f = 0.15), border = NA)
}
# Plot ATAC-seq track
par(new = TRUE)
plot(0.5*(melp_5th_brain_PAN$panStart + melp_5th_brain_PAN$panEnd), melp_5th_brain_PAN$score, type='l', xlim = c(start,end), ylim = c(0,200), ylab = "", yaxt = "n", lwd = 1, xlab = "", xaxt = "n", main = "", bty='none', col = "black")
# Add labels
mtext('ATAC-seq 5th instar Brain', side = 1, cex=0.8, padj = 0, las = 1, adj=1)
axis(2, at = seq(0,200, by=50), line = 1)
mtext('Score', side = 2, cex=0.8, line = 3)
## Plot H. melpomene Forewing
plot(NULL, xlim=c(start,end), ylim = c(0,200), axes=FALSE, ann=FALSE)
# Add some shading for unique blocks under the plot
for(e in 1: nrow(unique_melp)){
rect(unique_melp$startPos[e],0,unique_melp$endPos[e],200, col = adjustcolor('deepskyblue',alpha.f = 0.15), border = NA)
}
# Plot ATAC-seq track
par(new = TRUE)
plot(0.5*(melp_5th_FW_PAN$panStart + melp_5th_FW_PAN$panEnd), melp_5th_FW_PAN$score, type='l', xlim = c(start,end), ylim = c(0,200), ylab = "", yaxt = "n", lwd = 1, xlab = "", xaxt = "n", main = "", bty='none', col = "black")
# Add labels
mtext('ATAC-seq 5th instar FW', side = 1, cex=0.8, padj = 0, las = 1, adj=1)
axis(2, at = seq(0,200, by=50), line = 1)
mtext('Score', side = 2, cex=0.8, line = 3)
## Plot H. erato brain
plot(NULL, xlim=c(start,end), ylim = c(0,200), axes=FALSE, ann=FALSE)
# Add some shading for unique blocks under the plot
for(e in 1: nrow(unique_erato)){
rect(unique_erato$startPos[e],0,unique_erato$endPos[e],200, col = adjustcolor('mediumseagreen',alpha.f = 0.15), border = NA)
}
# Plot ATAC-seq track
par(new = TRUE)
plot(0.5*(erato_5th_brain_PAN$panStart + erato_5th_brain_PAN$panEnd), erato_5th_brain_PAN$score, type='l', xlim = c(start,end), ylim = c(0,200), ylab = "", yaxt = "n", lwd = 1, xlab = "", xaxt = "n", main = "", bty='none', col = "black")
# Add labels
mtext('ATAC-seq 5th instar Brain', side = 1, cex=0.8, padj = 0, las = 1, adj=1)
axis(2, at = seq(0,200, by=50), line = 1)
mtext('Score', side = 2, cex=0.8, line = 3)
## Plot H. erato FW
plot(NULL, xlim=c(start,end), ylim = c(0,200), axes=FALSE, ann=FALSE)
# Add some shading for unique blocks under the plot
for(e in 1: nrow(unique_erato)){
rect(unique_erato$startPos[e],0,unique_erato$endPos[e],200, col = adjustcolor('mediumseagreen',alpha.f = 0.15), border = NA)
}
# Plot ATAC-seq track
par(new = TRUE)
plot(0.5*(erato_5th_FW_PAN$panStart + erato_5th_FW_PAN$panEnd), erato_5th_FW_PAN$score, type='l', xlim = c(start,end), ylim = c(0,200), ylab = "", yaxt = "n", lwd = 1, xlab = "", xaxt = "n", main = "", bty='none', col = "black")
# Add labels
mtext('ATAC-seq 5th instar FW', side = 1, cex=0.8, padj = 0, las = 1, adj=1)
axis(2, at = seq(0,200, by=50), line = 1)
mtext('Score', side = 2, cex=0.8, line = 3)
Let's now do the same for the Minimap2 alignment and the TE annotations.
6.6. Map the Minimap2 alignment to the pan genome and plot
6.6.1. Create Minimap2 mapping files
# Read in the Minimap2 alignment file
miniMap_out <- read.table('Minimap2_out/Minimap_melp_erato.sam', header = FALSE, sep = '\t')
# Set the column names
colnames(miniMap_out) <- c('queryName', 'queryLength', 'queryStart', 'queryEnd',
'char',
'targetName', 'targetLength', 'targetStart', 'targetEnd',
'matchingBases', 'matchLength', 'matchQuality')
# Extract the start and end position of the Minimap2 alignments.
miniMap_target_start <- as.data.frame(miniMap_out$targetStart)
miniMap_target_end <- as.data.frame(miniMap_out$targetEnd)
miniMap_query_start <- as.data.frame(miniMap_out$queryStart)
miniMap_query_end <- as.data.frame(miniMap_out$queryEnd)
# Add the first row which specifies the transform direction.
colnames(miniMap_target_start) <- paste("2\tc")
colnames(miniMap_target_end) <- paste("2\tc")
colnames(miniMap_query_start) <- paste("1\tc")
colnames(miniMap_query_end) <- paste("1\tc")
# Write the objects to files.
write.table(miniMap_target_start, "miniMap_target_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(miniMap_target_end, "miniMap_target_end.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(miniMap_query_start, "miniMap_query_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(miniMap_query_end, "miniMap_query_end.toMap.txt", quote = FALSE, row.names = FALSE)
6.6.2. Transform the Minimap2 positions
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i Minimap2_out/miniMap_target_start.toMap.txt -n Minimap2_out/miniMap_target_start.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i Minimap2_out/miniMap_target_end.toMap.txt -n Minimap2_out/miniMap_target_end.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i Minimap2_out/miniMap_query_start.toMap.txt -n Minimap2_out/miniMap_query_start.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i Minimap2_out/miniMap_query_end.toMap.txt -n Minimap2_out/miniMap_query_end.pan
6.6.3. Read and plot the transformed Minimap2 positions
# Read in the Minimap2 alignment file
miniMap_out <- read.table('Minimap2_out/Minimap_melp_erato.sam', header = FALSE, sep = '\t')
# Set the column names
colnames(miniMap_out) <- c('queryName', 'queryLength', 'queryStart', 'queryEnd',
'char',
'targetName', 'targetLength', 'targetStart', 'targetEnd',
'matchingBases', 'matchLength', 'matchQuality')
# Extract the start and end position of the Minimap2 alignments.
miniMap_target_start <- as.data.frame(miniMap_out$targetStart)
miniMap_target_end <- as.data.frame(miniMap_out$targetEnd)
miniMap_query_start <- as.data.frame(miniMap_out$queryStart)
miniMap_query_end <- as.data.frame(miniMap_out$queryEnd)
# Add the first row which specifies the transform direction.
colnames(miniMap_target_start) <- paste("2\tc")
colnames(miniMap_target_end) <- paste("2\tc")
colnames(miniMap_query_start) <- paste("1\tc")
colnames(miniMap_query_end) <- paste("1\tc")
# Write the objects to files.
write.table(miniMap_target_start, "Minimap2_out/miniMap_target_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(miniMap_target_end, "Minimap2_out/miniMap_target_end.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(miniMap_query_start, "Minimap2_out/miniMap_query_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(miniMap_query_end, "Minimap2_out/miniMap_query_end.toMap.txt", quote = FALSE, row.names = FALSE)
# 6.6.2. Transform the Minimap2 positions
# 6.6.3. Read and plot the transformed Minimap2 positions
miniMap_target_panPos_start <- read.table('Minimap2_out/miniMap_target_start.pan.txt', header = TRUE, sep = '\t')
miniMap_target_panPos_end <- read.table('Minimap2_out/miniMap_target_end.pan.txt', header = TRUE, sep = '\t')
miniMap_query_panPos_start <- read.table('Minimap2_out/miniMap_query_start.pan.txt', header = TRUE, sep = '\t')
miniMap_query_panPos_end <- read.table('Minimap2_out/miniMap_query_end.pan.txt', header = TRUE, sep = '\t')
colnames(miniMap_target_panPos_start) <- c("targetStart", "targetStartPAN")
colnames(miniMap_target_panPos_end) <- c("targetEnd", "targetEndPAN")
colnames(miniMap_query_panPos_start) <- c("queryStart", "queryStartPAN")
colnames(miniMap_query_panPos_end) <- c("queryEnd", "queryEndPAN")
# Merge the old with the newly transformed positions. In contrast to the bedgraphs, I here merge using the old position in the Minimap2 file, because the order of the alignment positions might be out of order between the two genomes.
miniMap_outPAN <- merge(miniMap_out, miniMap_target_panPos_start, by = 'targetStart')
miniMap_outPAN <- merge(miniMap_outPAN, miniMap_target_panPos_end, by = 'targetEnd')
miniMap_outPAN <- merge(miniMap_outPAN, miniMap_query_panPos_start, by = 'queryStart')
miniMap_outPAN <- merge(miniMap_outPAN, miniMap_query_panPos_end, by = 'queryEnd')
# Plot the alignment.
plot(NULL, xlim = c(start, end), ylim = c(0,10), axes=F, ylab = '', xlab = '')
for(e in 1:nrow(miniMap_outPAN)){
if(miniMap_outPAN$targetStartPAN[e] > start-20000 & miniMap_outPAN$targetEndPAN[e] < end+20000 & miniMap_outPAN$queryStartPAN[e] > start-20000 & miniMap_outPAN$queryEndPAN[e] < end+20000){
# if(miniMap_outPAN$matchingBases[e]/miniMap_outPAN$matchLength[e] > 0.2){
polygon(x = c(miniMap_outPAN$targetStartPAN[e], miniMap_outPAN$targetEndPAN[e], miniMap_outPAN$queryEndPAN[e], miniMap_outPAN$queryStartPAN[e]),
y = c(10,10,0,0),
col = adjustcolor('black', alpha.f = miniMap_outPAN$matchingBases[e]/miniMap_outPAN$matchLength[e]), border = FALSE)
# }
}
}
6.7. Map the TEs to the pan genome and plot
6.7.1. Create TE mapping files
# Read in the TE files
TE_erato <- read.table('TEs/H_erato_1801_TE.txt', header = FALSE)[,c(2,5:7)]
TE_melp <- read.table('TEs/H_melp_18003_TE.txt', header = FALSE)[,c(2,5:7)]
# Set the column names
colnames(TE_erato) <- c('subs','scaf', 'start', 'end')
colnames(TE_melp) <- c('subs','scaf', 'start', 'end')
# The files include TEs outside of the interval we are interested in, so we can remove those.
TE_erato <- subset(TE_erato, TE_erato$end < 2000000)
TE_melp <- subset(TE_melp, TE_melp$end < 2000000)
# (optional) remove TEs with large number of substitutions compared to database
TE_erato <- subset(TE_erato, TE_erato$subs < 15)
TE_melp <- subset(TE_melp, TE_melp$subs < 15)
# Extract the start and end position of the TEs.
TE_erato_start <- as.data.frame(TE_erato[,3])
TE_erato_end <- as.data.frame(TE_erato[,4])
TE_melp_start <- as.data.frame(TE_melp[,3])
TE_melp_end <- as.data.frame(TE_melp[,4])
# Add the first row which specifies the transform direction.
colnames(TE_erato_start) <- paste("1\tc")
colnames(TE_erato_end) <- paste("1\tc")
colnames(TE_melp_start) <- paste("2\tc")
colnames(TE_melp_end) <- paste("2\tc")
# Write the objects to files.
write.table(TE_erato_start, "TE_erato_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(TE_erato_end, "TE_erato_end.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(TE_melp_start, "TE_melp_start.toMap.txt", quote = FALSE, row.names = FALSE)
write.table(TE_melp_end, "TE_melp_end.toMap.txt", quote = FALSE, row.names = FALSE)
6.7.2. Transform the TE positions
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i TEs/TE_erato_start.toMap.txt -n TEs/TE_erato_start.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i TEs/TE_erato_end.toMap.txt -n TEs/TE_erato_end.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i TEs/TE_melp_start.toMap.txt -n TEs/TE_melp_start.pan
seq-seq-pan map -c seq-seq-pan_out/SeqSeqPan_erato_melp_optix_consensus.fasta -p ./ -i TEs/TE_melp_end.toMap.txt -n TEs/TE_melp_end.pan
6.7.3. Read and plot the transformed TE positions
TE_erato_panPos_start <- read.table('TEs/TE_erato_start.pan.txt', header = TRUE, sep = '\t')
TE_erato_panPos_end <- read.table('TEs/TE_erato_end.pan.txt', header = TRUE, sep = '\t')
TE_melp_panPos_start <- read.table('TEs/TE_melp_start.pan.txt', header = TRUE, sep = '\t')
TE_melp_panPos_end <- read.table('TEs/TE_melp_end.pan.txt', header = TRUE, sep = '\t')
TE_erato_PAN <- cbind(as.data.frame(TE_erato_panPos_start[,2]),as.data.frame(TE_erato_panPos_end[,2]))
TE_melp_PAN <- cbind(as.data.frame(TE_melp_panPos_start[,2]),as.data.frame(TE_melp_panPos_end[,2]))
colnames(TE_erato_PAN) <- c('startPAN', 'endPAN')
colnames(TE_melp_PAN) <- c('startPAN', 'endPAN')
# Remove potential erroneous mappings which can be identified as start/end positions too far apart.
TE_erato_PAN <- subset(TE_erato_PAN, abs(TE_erato_PAN$endPAN - TE_erato_PAN$startPAN) < 10000)
TE_melp_PAN <- subset(TE_melp_PAN, abs(TE_melp_PAN$endPAN - TE_melp_PAN$startPAN) < 10000)
# Add the TEs to the current plot as rectangles
for(e in 1:nrow(TE_erato_PAN)){
rect(TE_erato_PAN$startPAN[e],-1,TE_erato_PAN$endPAN[e],0, col = 'orange', border = NA)
}
for(e in 1:nrow(TE_melp_PAN)){
rect(TE_melp_PAN$startPAN[e],10,TE_melp_PAN$endPAN[e],11, col = 'orange', border = NA)
}
# Add some labels
mtext('TE', side = 1, cex=0.5, padj = 0, las = 1, adj=1, col = 'orange')
mtext('Minimap2 alignment', side = 2, cex=0.8, padj = -1, col = 'black')
Pfieuw. We're done! You should now see the figure that was at the top of this tutorial.