In these tutorials I guide you through code for a Minimap2 genome alignment and a seq-seq-pan pan-genome alignment with visualizations in R. The tutorial uses an example of two haplotypes of the optix locus in Heliconius butterflies and will build these figures:
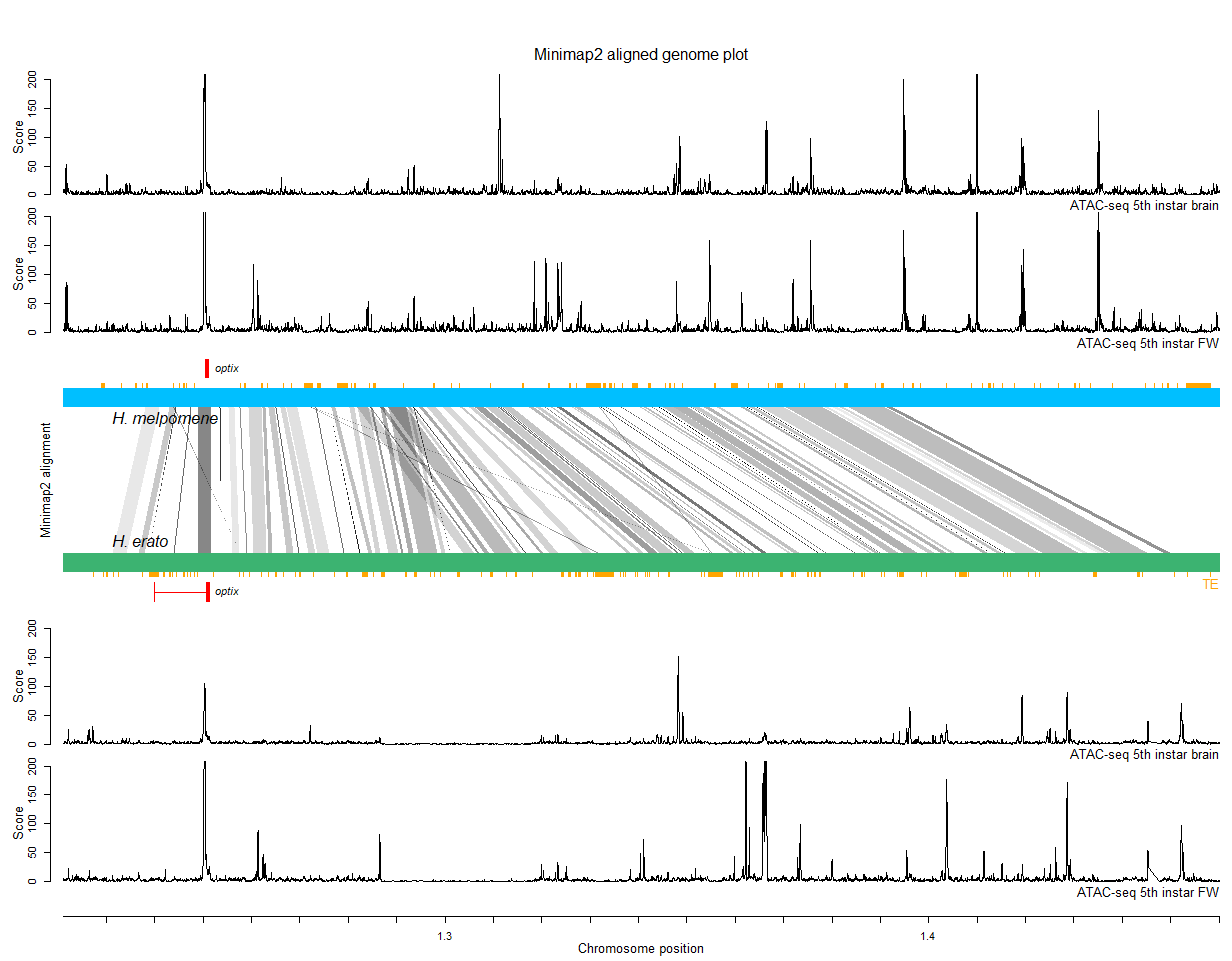
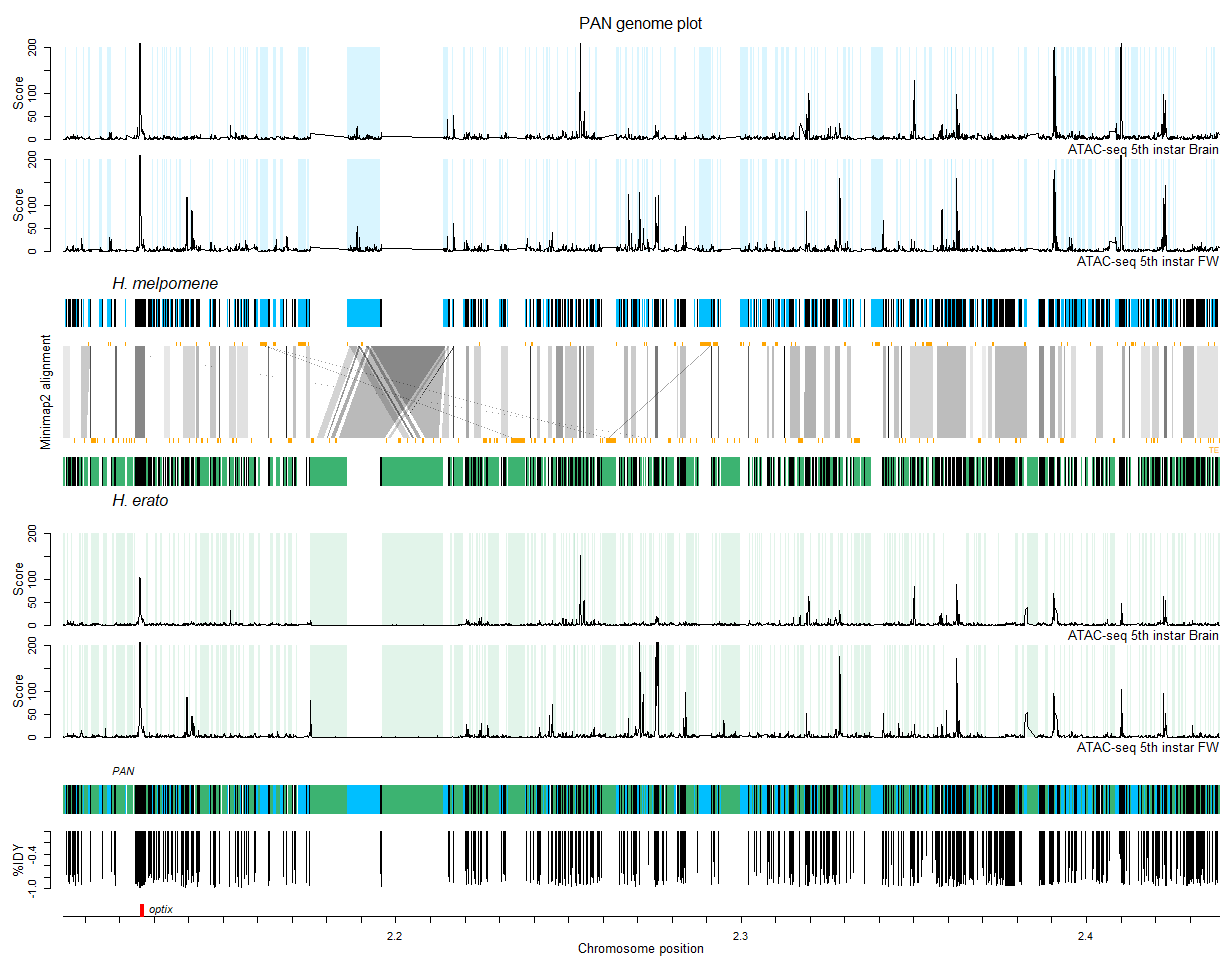
Similar code has been used to create figures in:
-
Van Belleghem SM, Ruggieri AA, Concha C, Livraghi L, Hebberecht L, Santiago Rivera E, Ogilvie JG, Hanly JJ, Warren IA, Planas S, Ortiz-Ruiz Y, Reed R, Lewis JJ, Jiggins CD, Counterman BA, McMillan WO, Papa R (2023). High level of novelty under the hood of convergent evolution. Science 379, 1043–1049.
-
Ruggieri AA, Livraghi L, Lewis JJ, Evans E, Cicconardi F, Hebberecht L, Montgomery SM, Ghezzi A, Rodriguez-Martinez JA, Jiggins CD, McMillan WO, Counterman BA, Papa R & Van Belleghem SM (2022). A butterfly pan-genome reveals that a large amount of structural variation underlies the evolution of chromatin accessibility. Genome Research 32: 1862-1875.
-
Livraghi L, Hanly JJ, Van Belleghem SM, Montejo-Kovacevich G, van der Heijden ESM, Loh LS, Ren A, Warren IA, Lewis JJ, Concha C, López LH, Wright C, Walker JM, Foley J, Goldberg Z, Arenas-Castro H, Brenes LR, Salazar C, Perry M, Papa R, Martin A, McMillan WO & Jiggins CD (2021). Cortex cis-regulatory switches establish scale colour identity and pattern diversity in Heliconius. eLife e68549.
-
Moest M, Van Belleghem SM, James J, Salazar C, Martin SH, Barker S, Moreira GRP, Merot C, Joron M, Nadeau N, Steiner F & Jiggins CD (2020). Classic and introgressed selective sweeps shape mimicry loci across a butterfly adaptive radiation. PloS Biology 18: e3000597.